Small compound screens are the classical application of high-throughput screening technology. Here, a large number of chemical compounds or drugs are tested for a desirable cellular response. In order to analyse this type of screen, select “Small compounds” on the first page of HiTSeekR.
See gene silencing
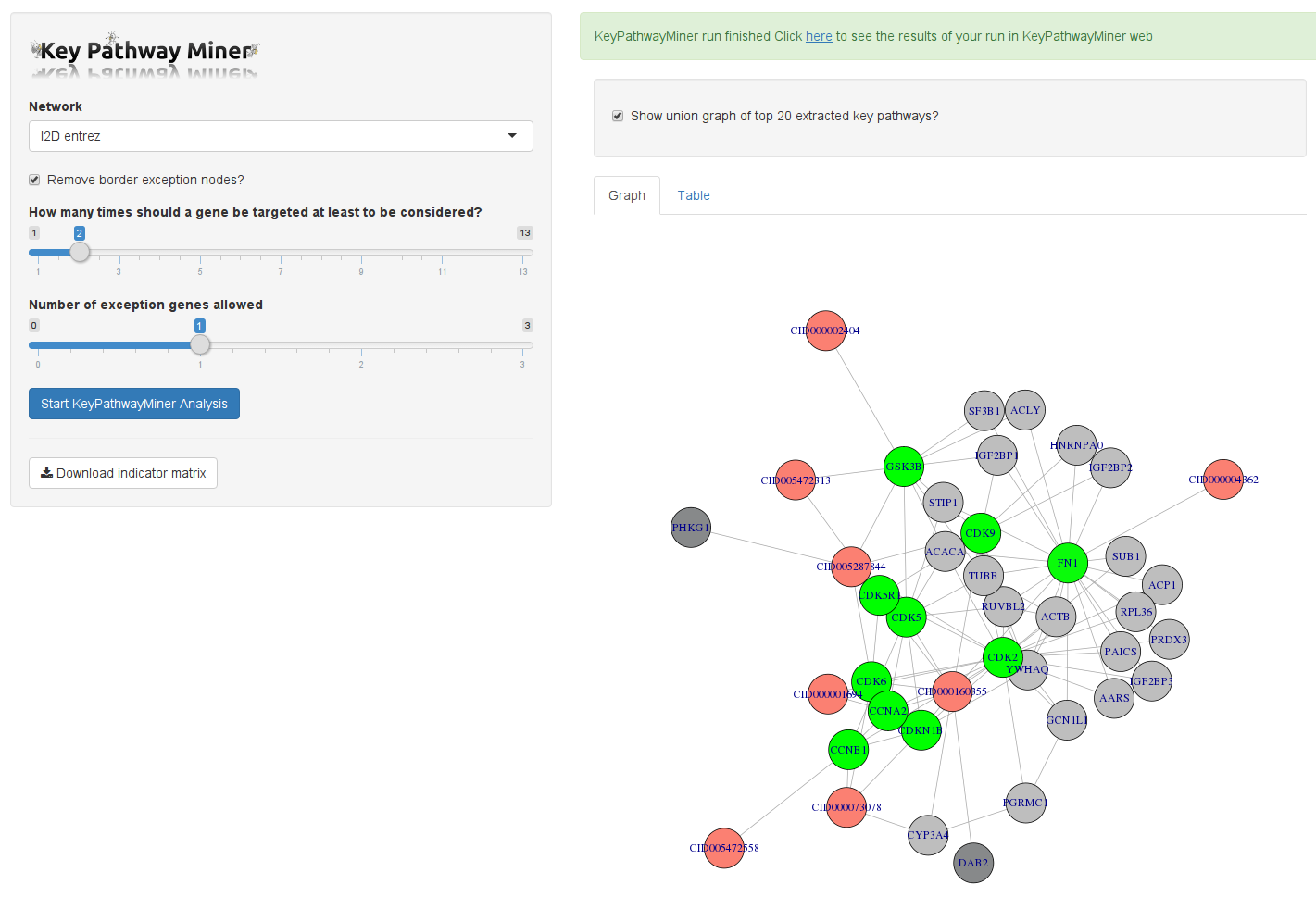
HiTSeekR is integrated with the STITCH database, which allows for identifying target genes via PubChem compound ids (CIDs) previously mapped to the hit compounds. The list of target genes can then be used in gene set and network enrichment analysis.
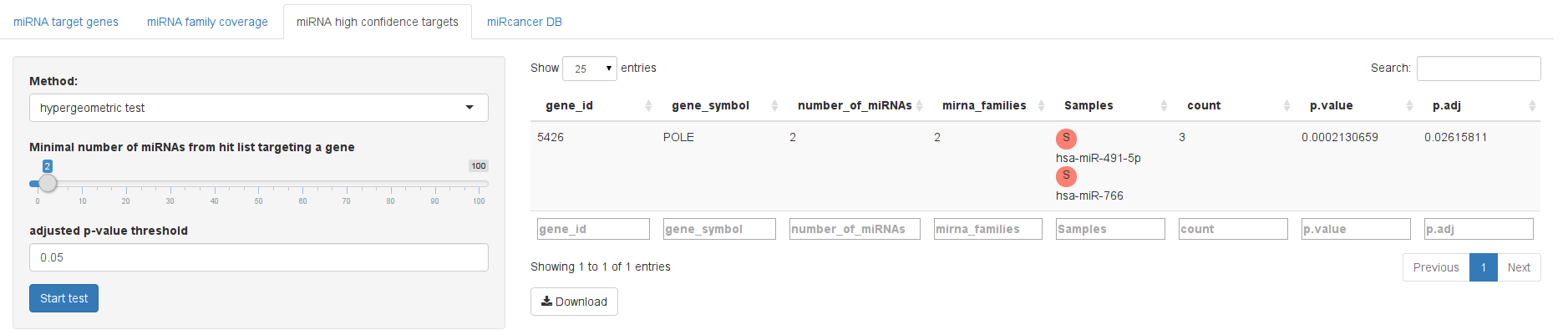
Same as in gene silencing but without support for gene set enrichment analysis.
In addition to showing the extracted subnetworks, HiTSeekR will also include promotors and suppressors connected to these genes. In addition to selecting the number of exception genes that KeyPathwayMiner will be allowed to add, there is now a second parameter that enables the user to influence when a gene is considered active. This threshold is the minimum number of different compounds that needs to target a given gene for it to be considered.