MicroRNAs are a class of RNAs consisting of small approximately 21-25 nucleotide long non-coding RNA molecules. They are synthesized by cells in a complex multi-step process to mature miRNAs, which have the ability to bind to mRNA through partial complementarity. Their main function is the down-regulation of gene expression and in contrast to, for example, siRNAs, they can target various genes, making them key suspects in the global regulation of gene expression. miRNAs have been implicated in various diseases and are thus of high interest for systematic research in high-throughput settings.
There are two types of miRNA screens:
Both are treated the same way in the analysis in HiTSeekR, which begins with selecting the miRNA screen type.
The processing of miRNA screening data is the same as for gene silencing screens. Once hits have been identified, however, the downstream analysis is different due to the nature of miRNAs, which operate through regulating expression of a large number of genes.
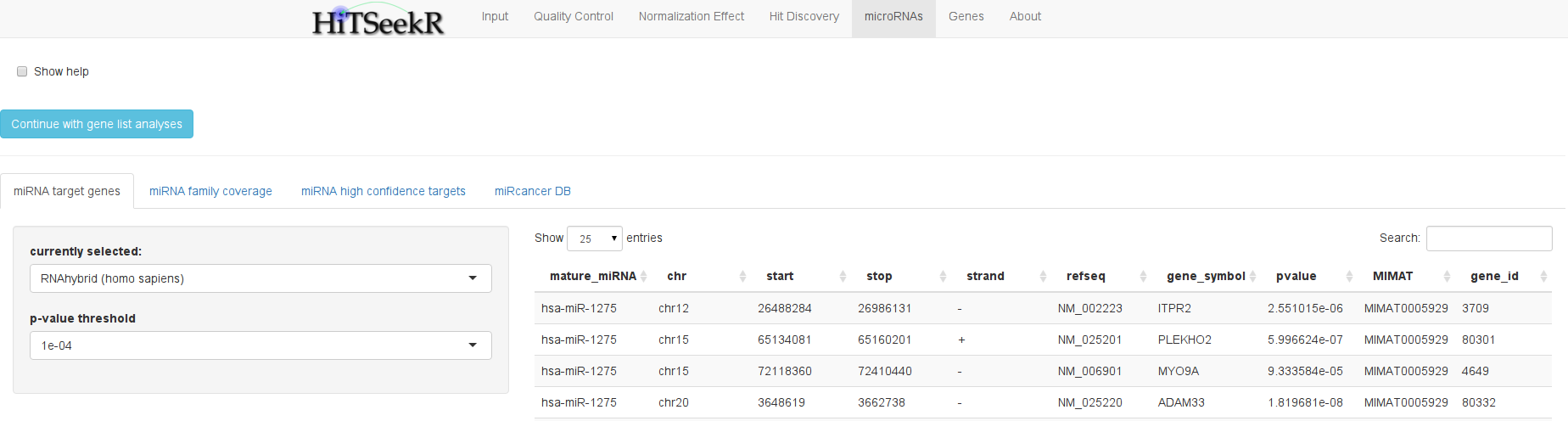
Since microRNAs themselves do not offer much information on their biological role it is crucial to learn which genes they regulate. To this end, a large number of predictive algorithms has been proposed. The most common ones are integrated here as databases. In addition, there are databases of experimentally validated miRNA targets like mirtarbase and tarbase. These provide only limited insights into microRNA targets, but on the other hand, they do not suffer from excessive false positive rates as the predictive methods do.
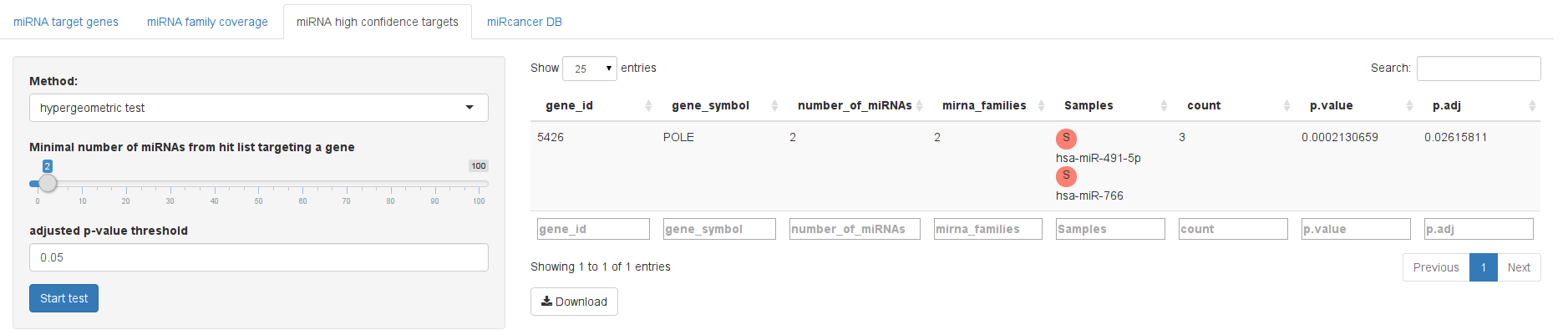
To deal with the relatively high false positive rate of miRNA prediction methods we sought a method to extract those genes that we believe are targeted with high confidence. For this, we wanted to exploit the fact that some miRNAs might cause a phenotype through regulating the same genes. If a gene is thus targeted by multiple hits of a screen this could be relevant. However, depending on the database and chosen thresholds, a gene can be targeted by hundreds of miRNAs. In other words, observing that gene X is targetd by Y different microRNAs active in a screen might be due to chance and a random selection of miRNAs might yield the same result. To deal with this effect, we computed p-values based on hypergeometric tests for RNAhybrid and based on permutation tests for all other databases.
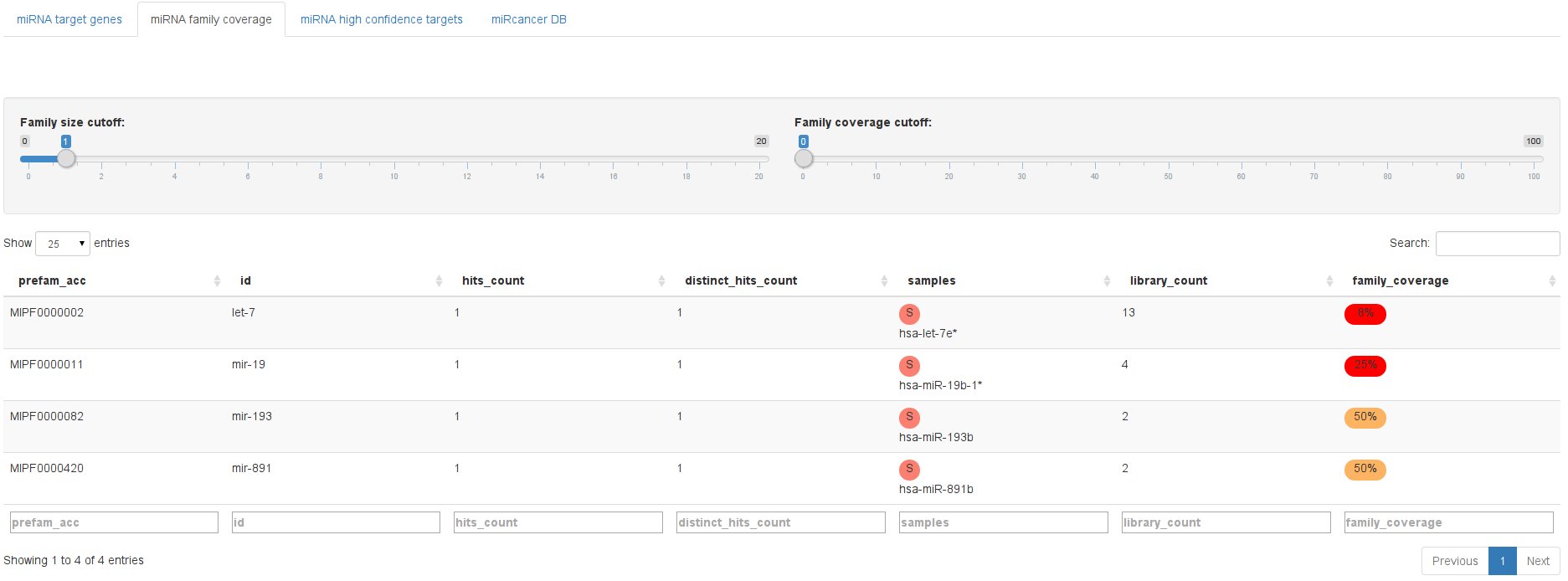
MicroRNAs are grouped into families based on their seed sequences. Since the seed sequence has the largest impact on target gene prediction, it can be assumed that several members of the same miRNA family might show the same phenotype. In contrast, if only a single member of a family shows an effect, this might very well be an off-target effect. We thus calculate the percentage of family members that appear to be active and show them here.
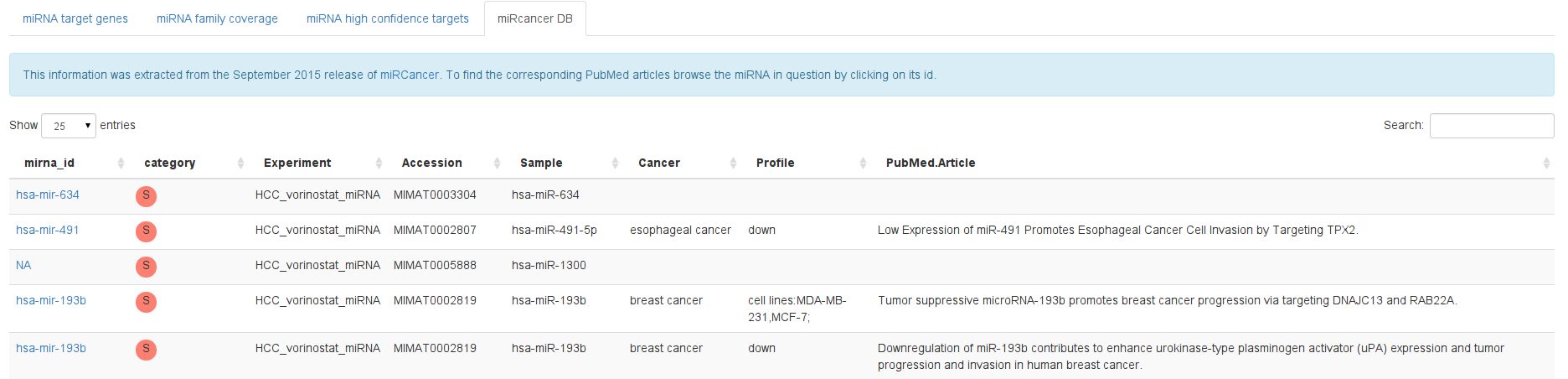
Deregulated miRNAs have been implicated in many diseases. miRcancer DB is a literature database that links miRNA identifiers to related publications in PubMed, together with a short description.
Same as in gene silencing but without support for gene set enrichment analysis.
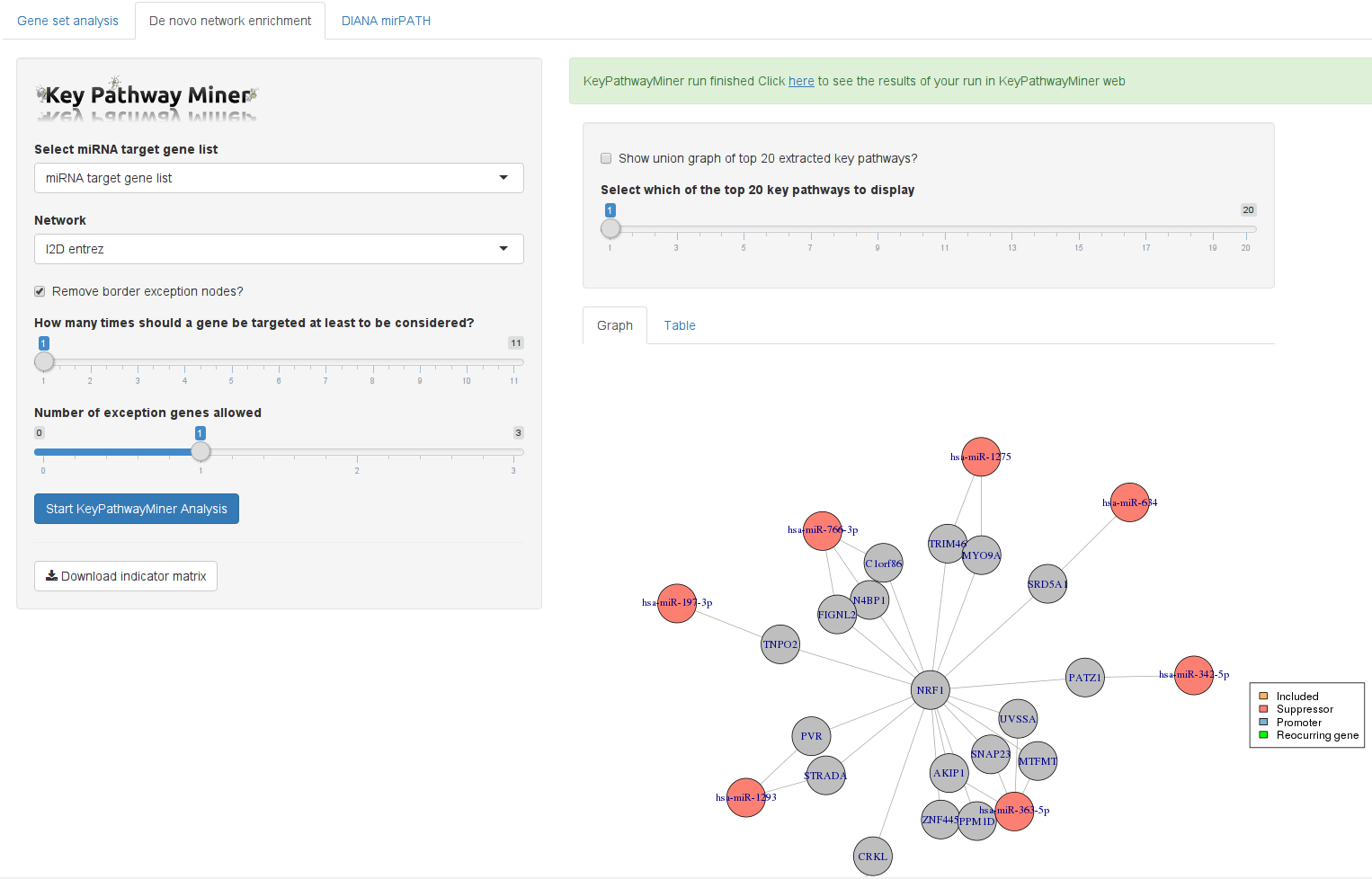
In addition to showing the extracted subnetworks, HiTSeekR will also include promoting and suppressing miRNA samples and connect them appropriately. In addition to selecting the number of exception genes that KeyPathwayMiner will be allowed to add, there is now a second parameter that enables the user to influence when a gene is considered active. This threshold is the minimum number of different miRNAs that needs to target a given gene for it to be considered.
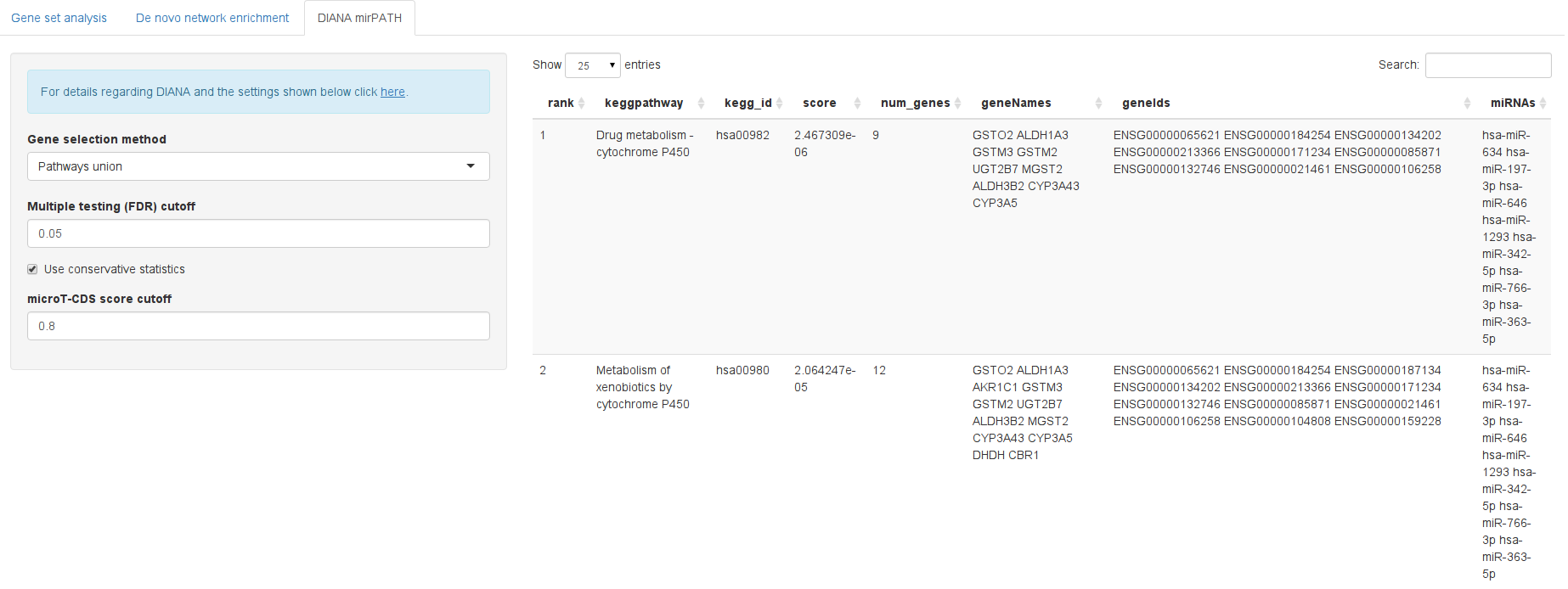
DIANA mirPATH is a tool dedicated to identify affected KEGG pathways in various ways, i.e. by building intersections or unions of targeted genes or targeted pathways. For details please refer to http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index Since mirPATH offers a webservice it could easily be integrated into HiTSeekR. Please note that long processing times may occur when using this feature due to external network traffic: